
Julienne Carstens, Ph.D., an assistant professor in the Division of Hematology and Oncology and associate scientist in the O’Neal Comprehensive Cancer Center, spends a lot of her time in sketchy neighborhoods.

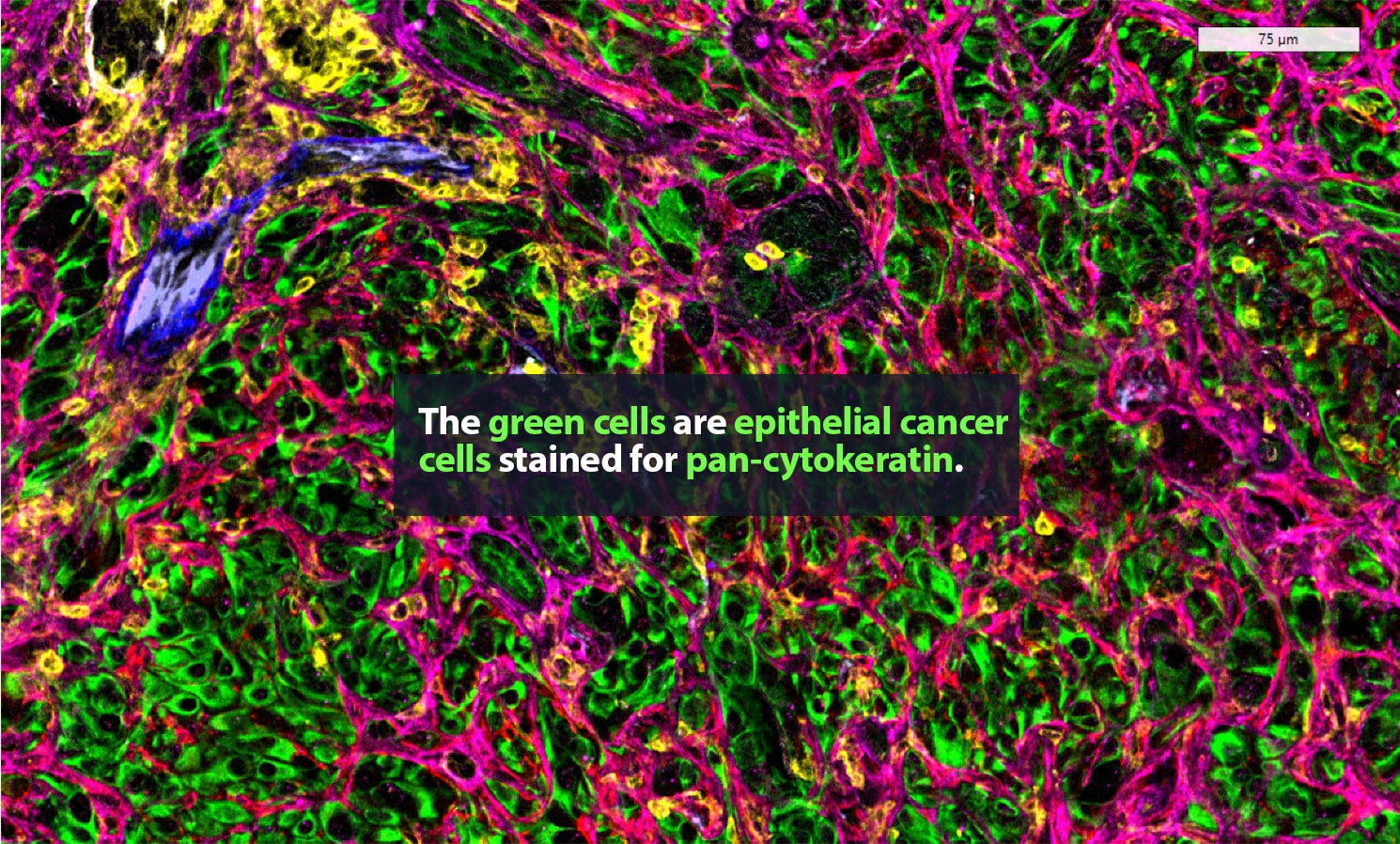
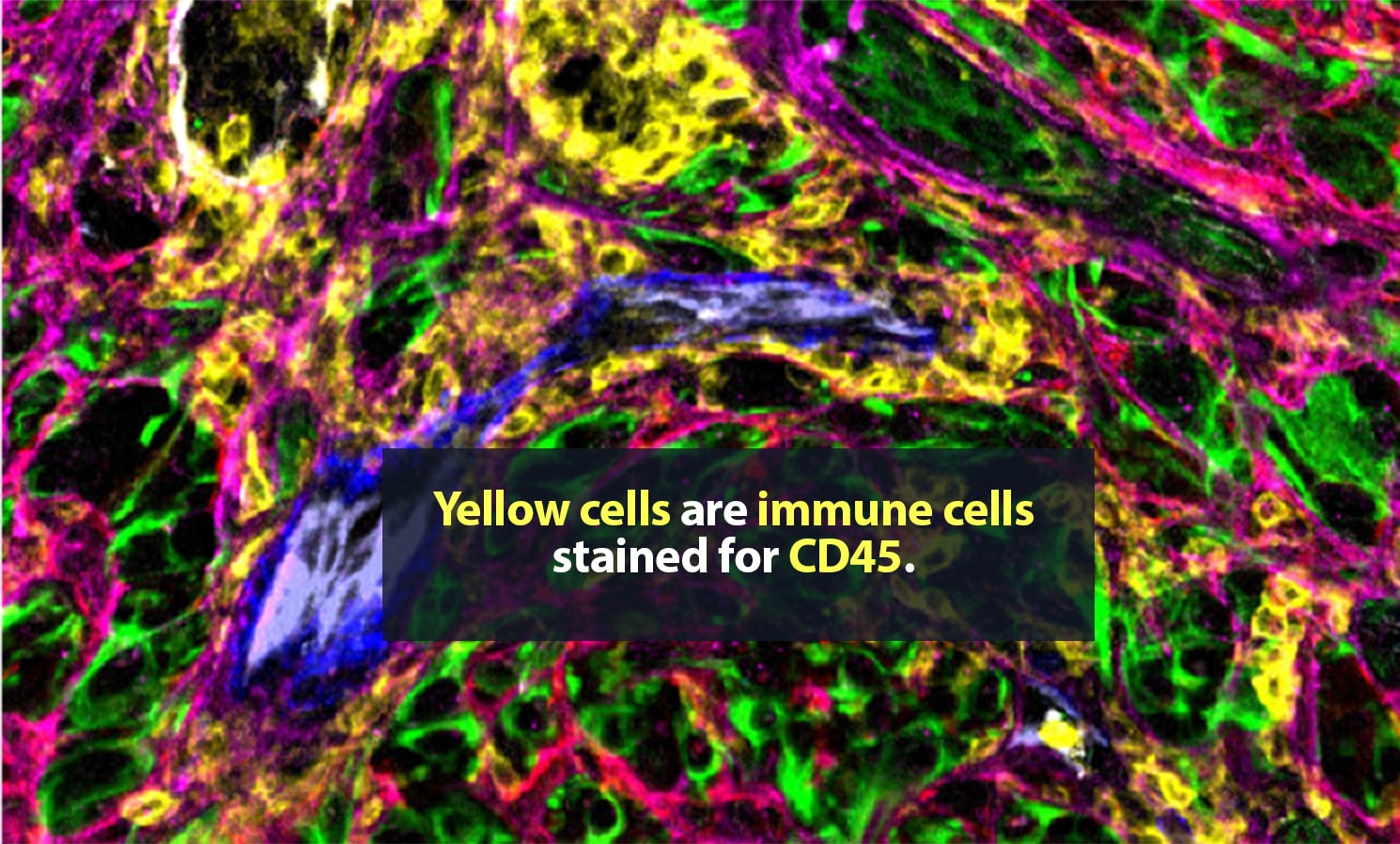
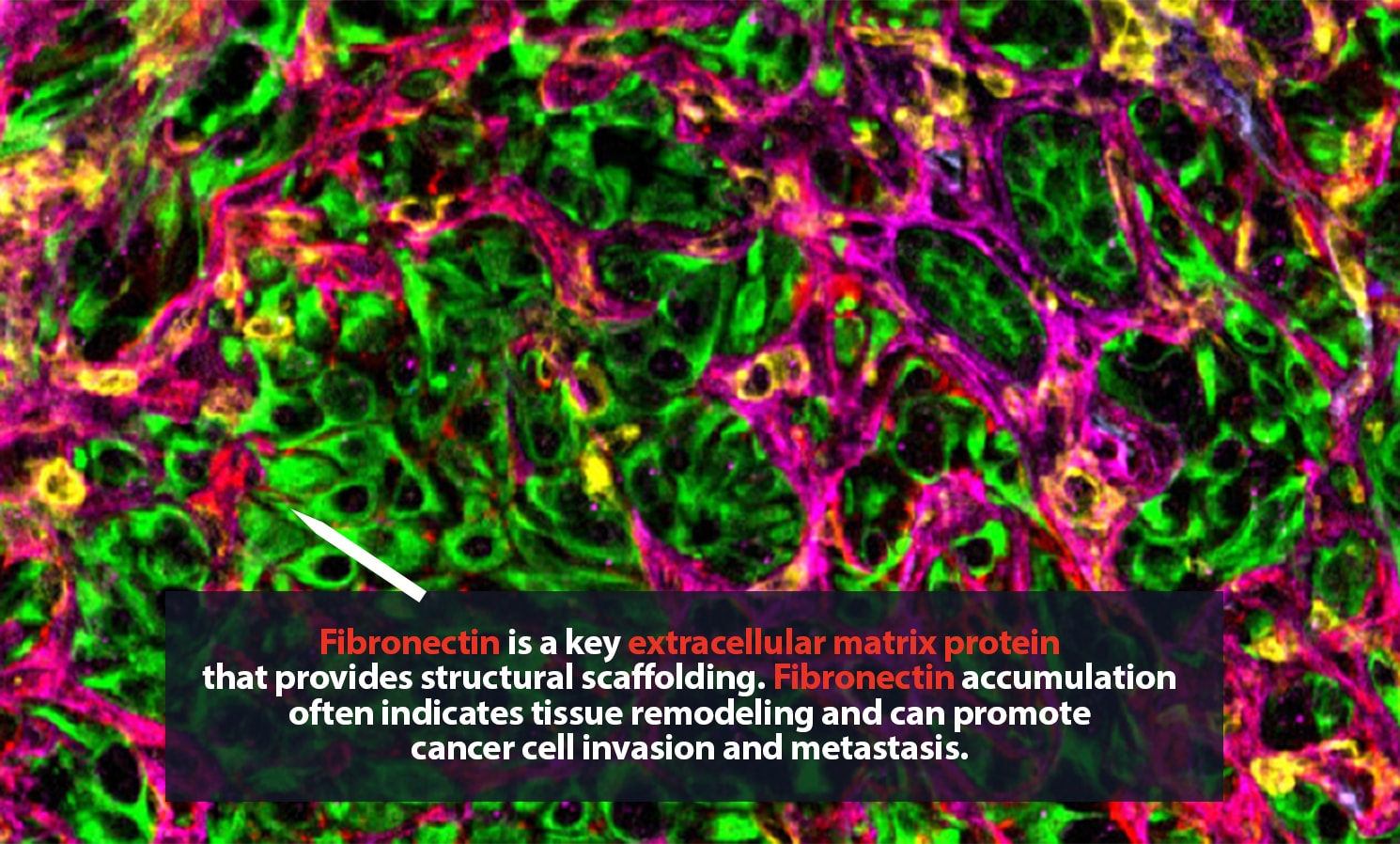
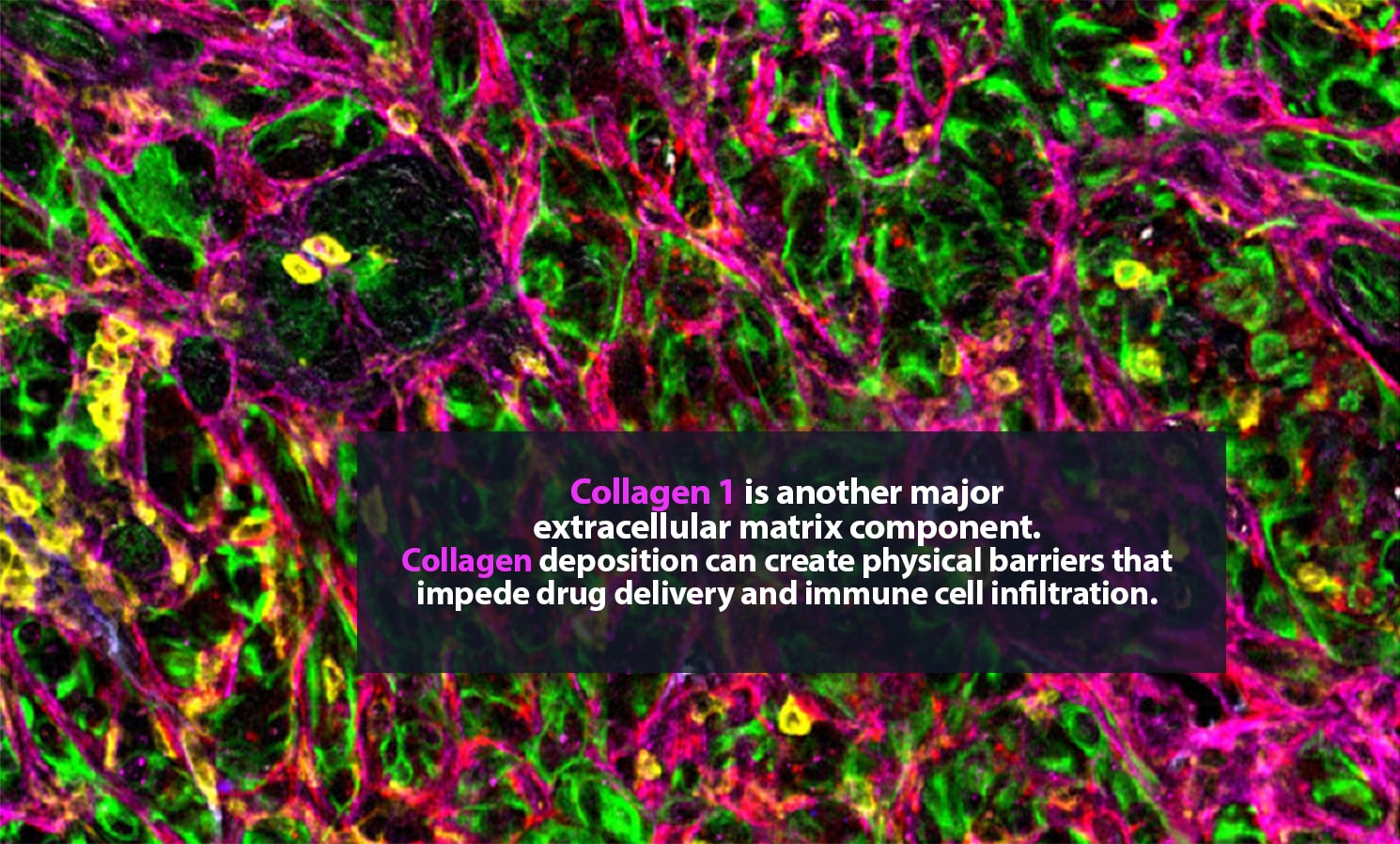
Carstens is in no personal danger. These spaces, samples from metastatic pancreatic cancer tumors, are typically only a few millimeters wide. Still, they can hold tens of thousands of cells, many of them single-mindedly bent on killing their host body. The environment around a tumor is a complex, fascinating place. Cancer cells are here, of course. So are many ordinary cells. Some of the rest are part of the body’s immune response to cancer. Then there are cells that, though they are not cancerous, are actively helping the tumor thrive.
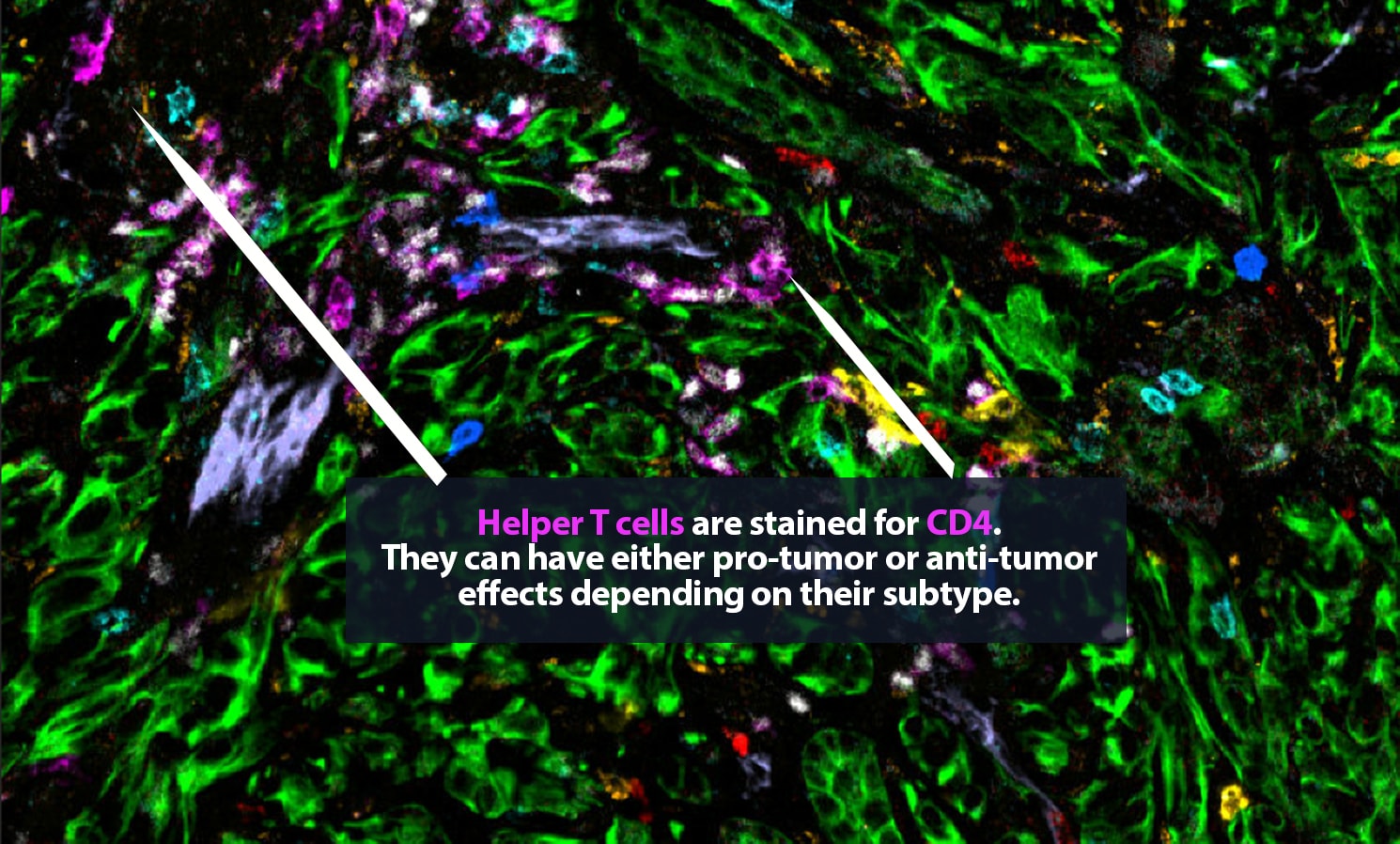
But which ones are which? Under a microscope, it can be difficult to tell a CD8+ cytotoxic T cell, a crucial part of the body’s defenses, from a regulatory T cell, which may have been co-opted by a tumor and is actively hampering that defense. Certainly, it is very hard to distinguish a vigorous CD8+ cell that is pumping out tumor-killing granzyme molecules from an exhausted CD8+ cell that is, essentially, doing nothing.
 Carstens lab member Jace Baines, researcher in the Department of Medicine.
Carstens lab member Jace Baines, researcher in the Department of Medicine.
Working with the same tumor slices that pathologists use to diagnose and guide treatment of patients, however, Carstens can create intricate, colorful maps of the microenvironment surrounding a tumor. She specializes in pancreatic cancer, a deadly and highly metastatic disease.
Her lab adds specific antibodies for every cell or protein of interest; each antibody emits a unique spectrum of light. “It is like one of those paint-by-numbers pictures, where each color is a different antibody,” Carstens said. Combining that information into a single image, she can generate a highly detailed picture of what is happening in the tumor neighborhood — who is there and what they are doing. She is a pioneer in the application of spatial analysis to cancer. “To better understand what certain cells are doing, it is important to know not just what cell type is present, but where it is,” she said.
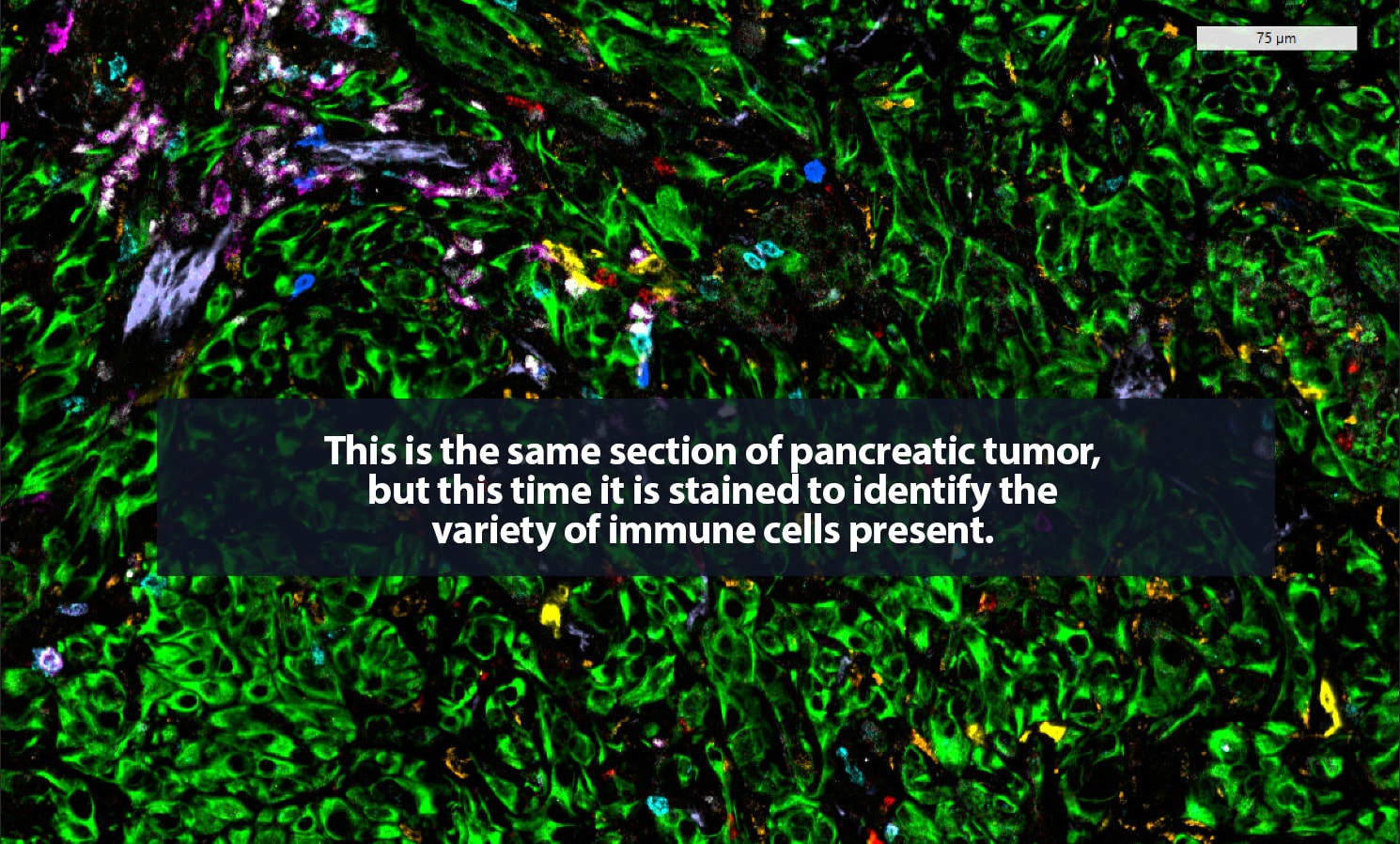
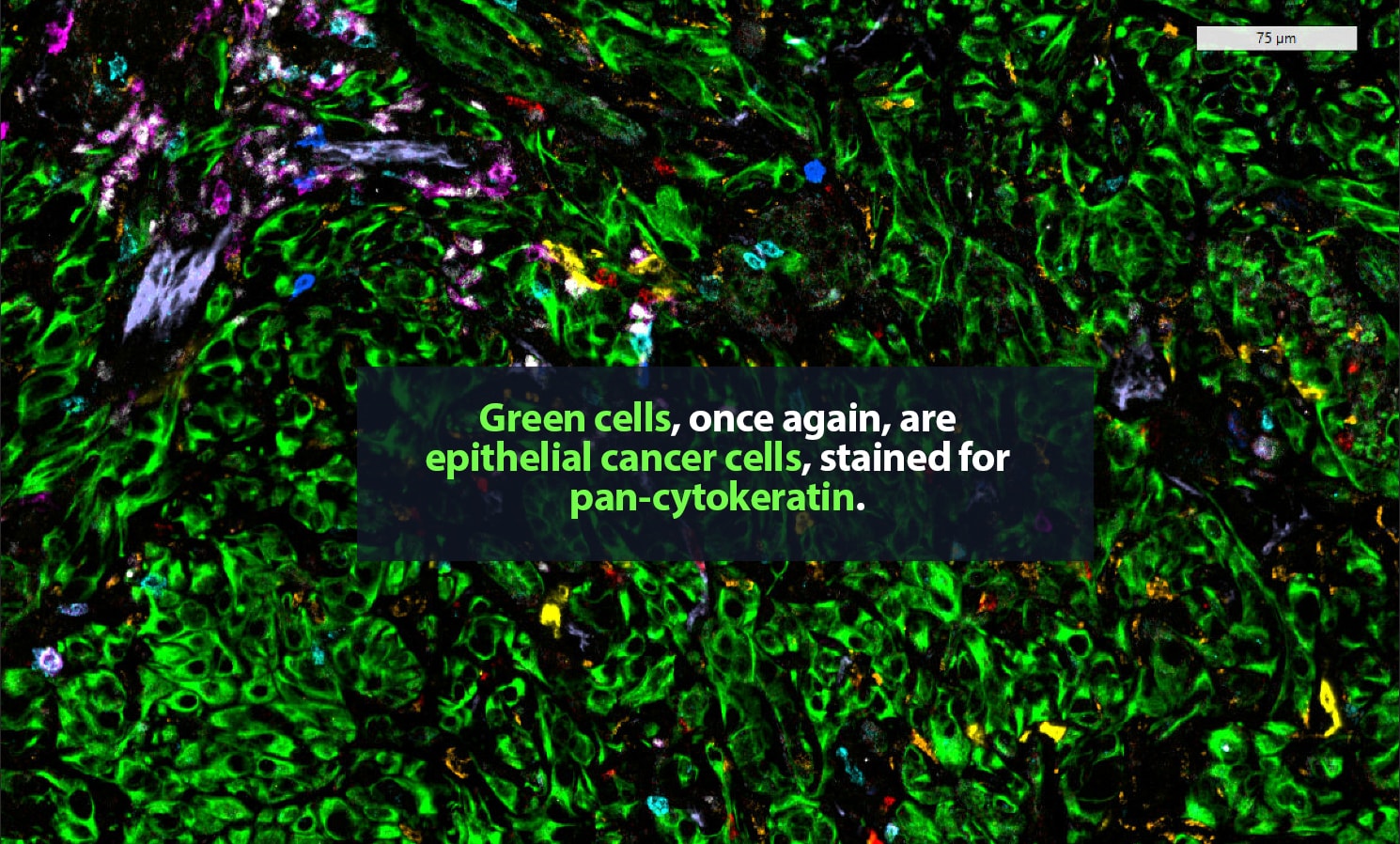
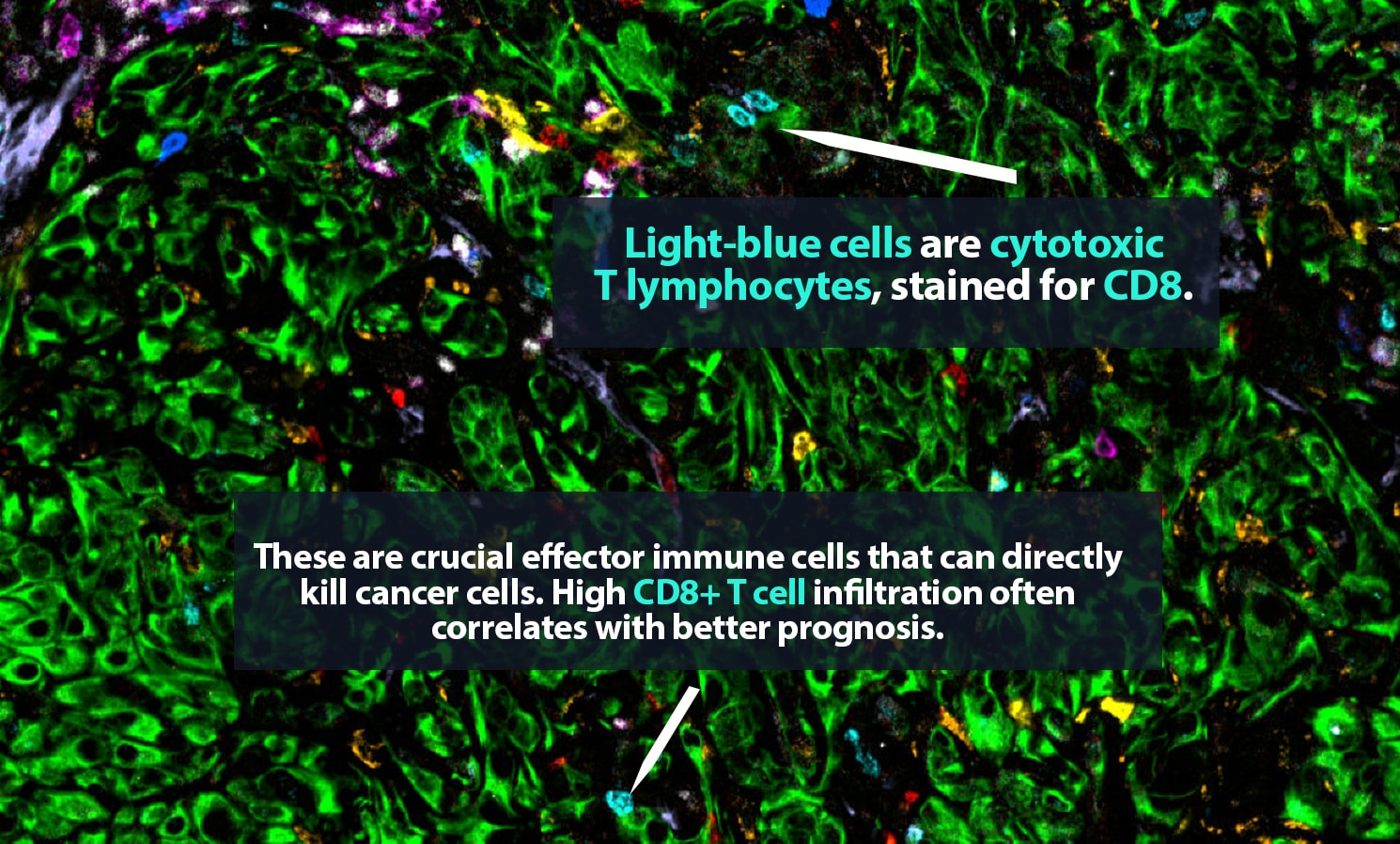
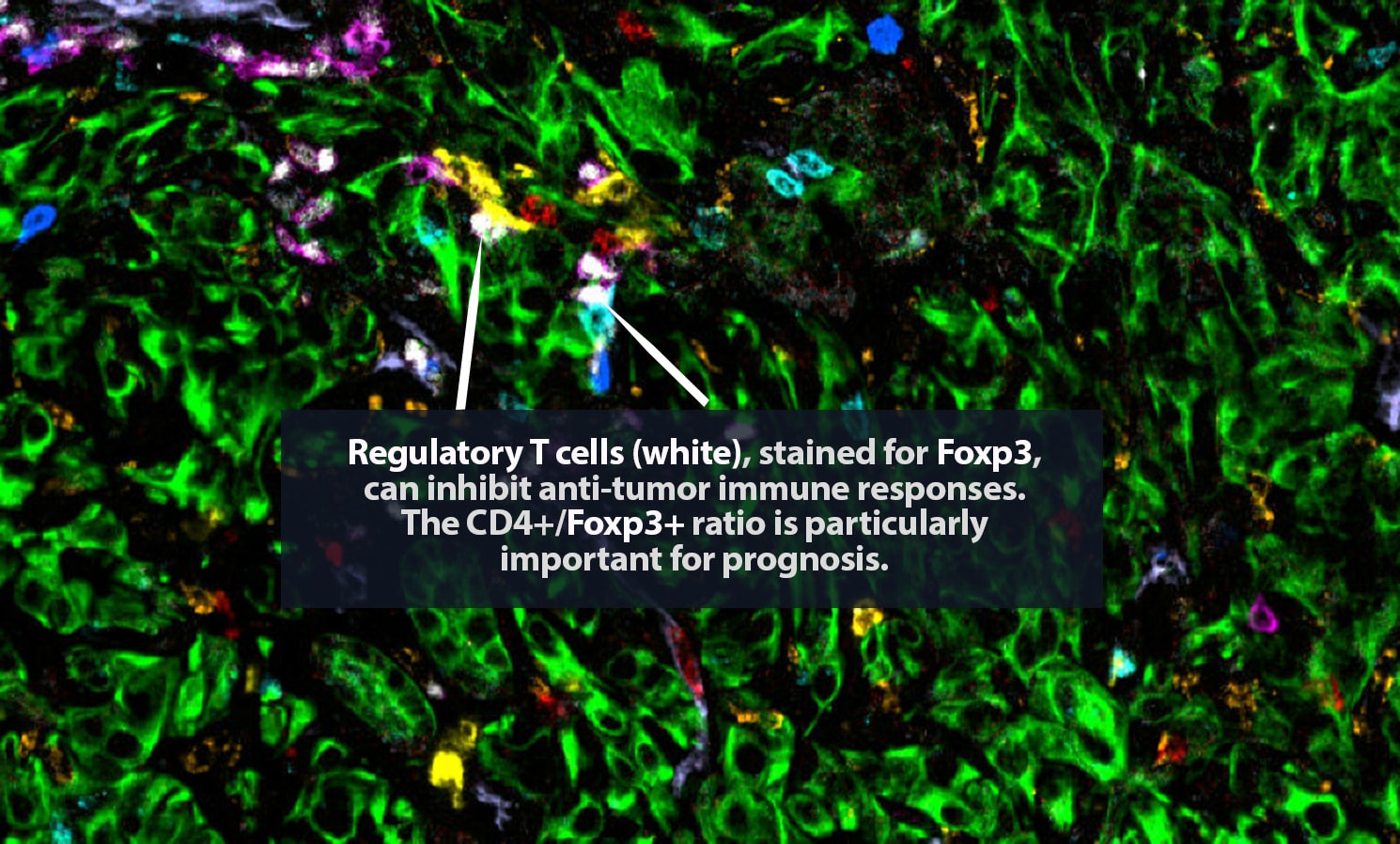
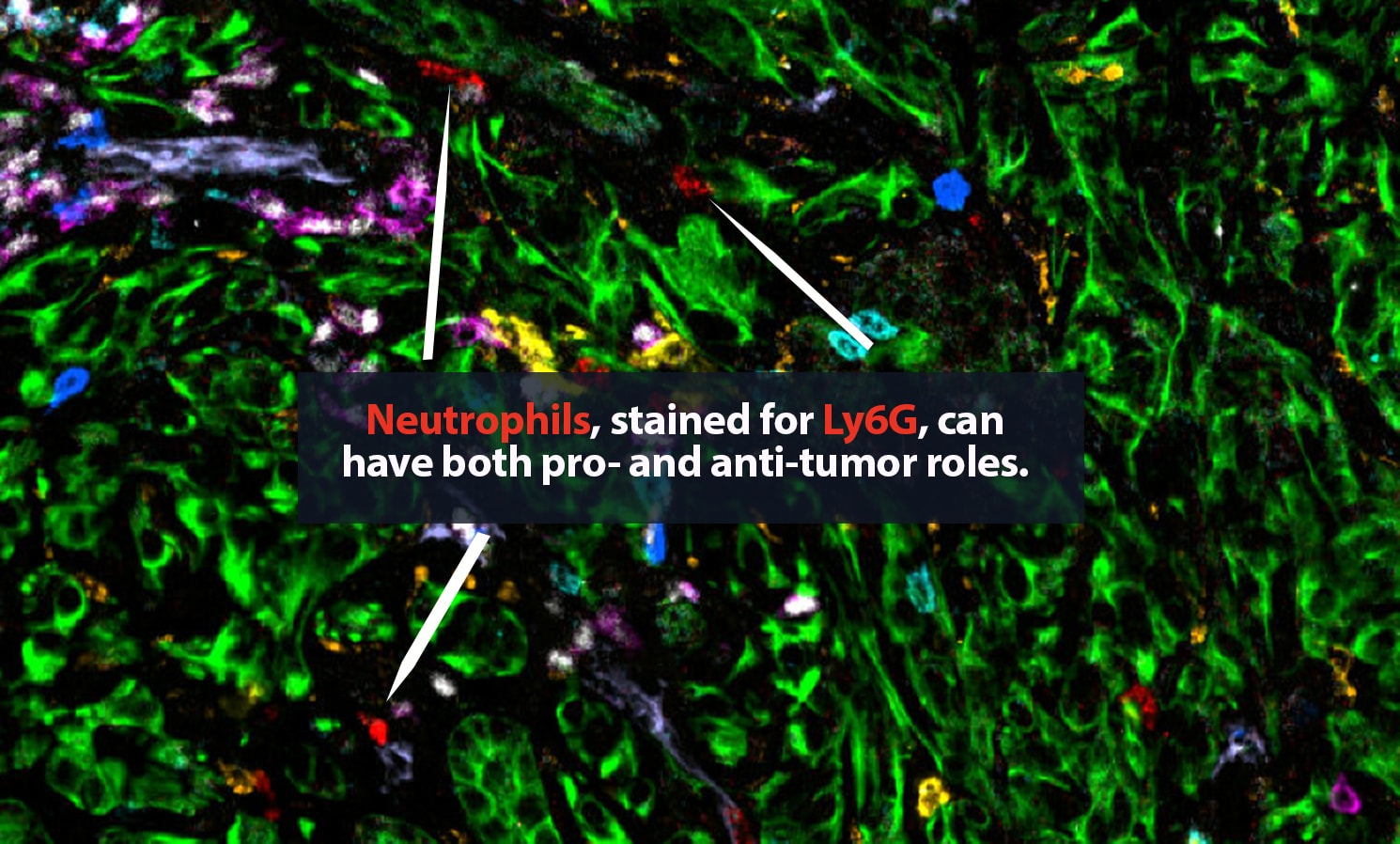
In 2017, Carstens, her collaborator Pedro Correa de Sampaio, Ph.D., and colleagues were the first to demonstrate that pancreatic cancer patients who have groups of cytotoxic CD8+ T cells located within 20 micrometers of their cancer cells — close enough to kill — are more likely to survive longer than patients who do not.
The state of the art in this area is advancing rapidly. For that 2017 paper, Carstens and Correa de Sampaio were the first to track eight different cell types in the tumor microenvironment. That result took a year and a half of work, including five full days of laborious pipetting and two days of microscopic imaging on Carstens’ part. Today, her lab can track 48-plus different proteins and the cell types they label at a time and complete the procedure in two days. “It is amazing how quickly the field has moved forward with multiplexing,” Carstens said.
 Carstens lab member Christina Larson, doctoral graduate student in the Cancer Biology Theme.
Carstens lab member Christina Larson, doctoral graduate student in the Cancer Biology Theme.
Now, the Carstens lab is exploring cutting-edge techniques to track how cell populations change over time — essentially turning her paintings into animated comic strips. One technique is to add unique genetic barcodes to cells in mouse models of cancer. By imaging samples over time, researchers can reconstruct the movements of specific cells as they divide and spread in the tumor neighborhood and travel around the body.
“We have started to learn that it is not just where the cancer cells come from that matters in metastasis — not just pancreatic cancer that is now in the liver — but some of my studies have shown that how they got there matters,” Carstens said. Researchers once thought that the epithelial to mesenchymal transition — from a stationary to a highly mobile form — was essential to metastasis. Carstens showed that locking cells into an epithelial state did not keep them from metastasizing. But it did greatly increase the odds that the cells would metastasize to the liver instead of the lungs, which is the most common and deadly metastatic site for pancreatic cancer patients. That suggests the possibility of predicting where to look for metastases — and ways of stopping the cells that do spread.
By adding temporal analyses to spatial analyses, Carstens believes that researchers can make progress in answering some of cancer’s thorniest questions. “Why does cancer metastasize to some places in some people but not others?” she said. “What causes cancer to take off and grow in spite of growth-killing chemotherapies? Could we influence how that cancer is going to respond?”
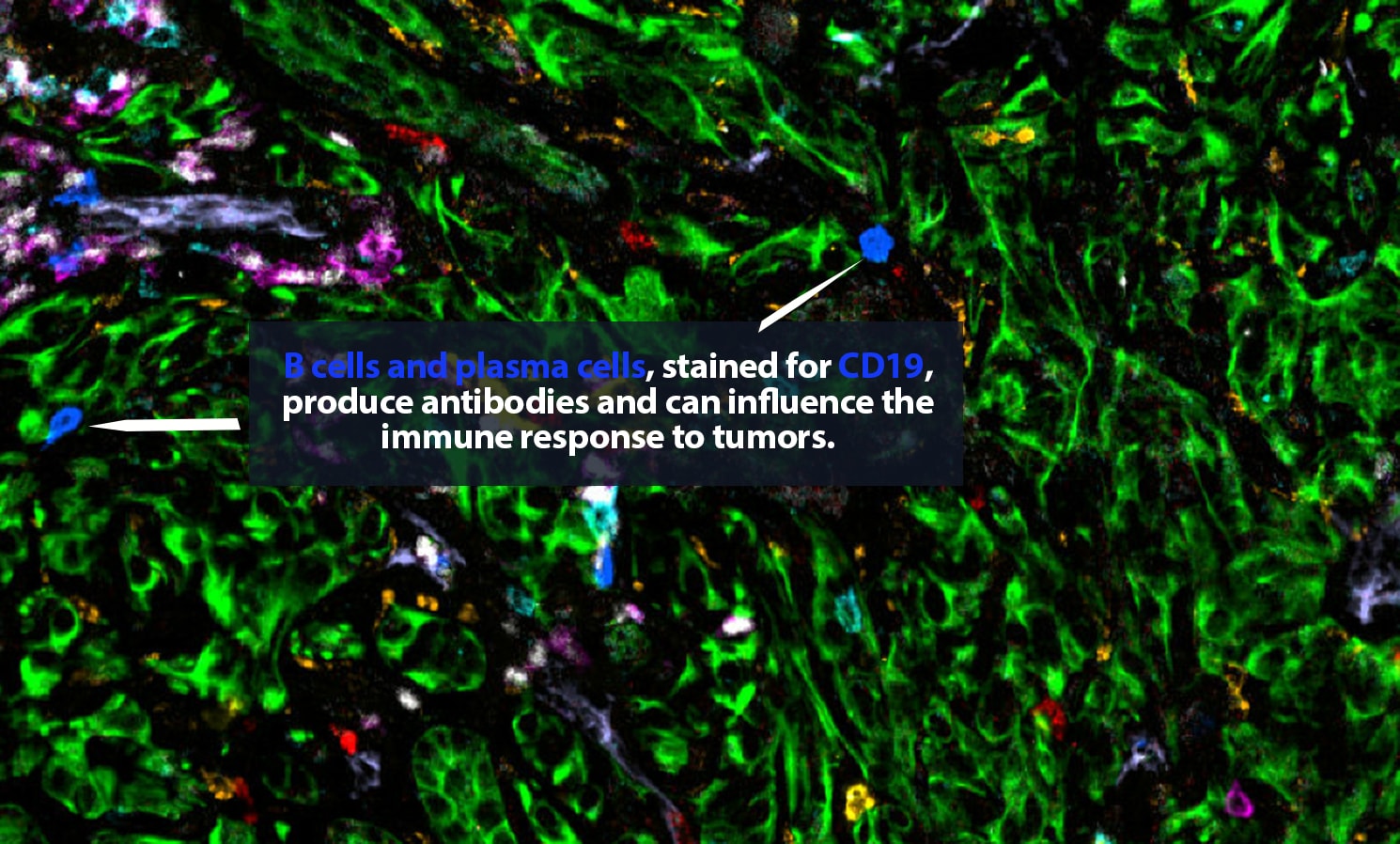
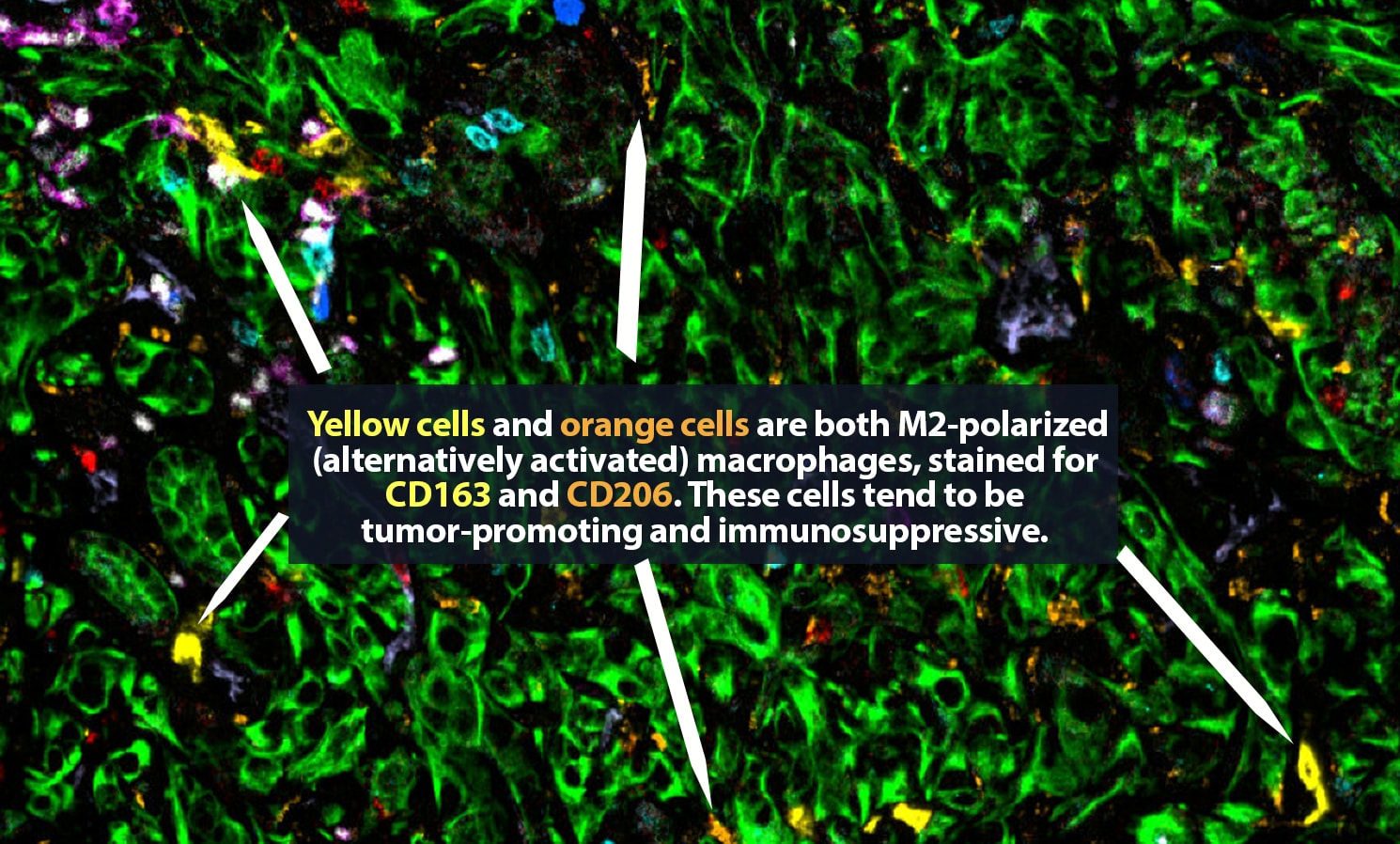
Traditional biopsies and surgical samples cannot answer those questions. A typical biopsy, Carstens explains, captures only a single snapshot in time. “We take this three-dimensional tumor and take one tiny two-dimensional section, and we draw a lot of conclusions from that,” she said. Based on that biopsy, the patient will be given one or more treatments. But within a few weeks, the tumor is fundamentally different. “It may have grown; it is under evolutionary selection pressure in which the cells responsive to chemo have died off and the resistant cells have grown. We want to track how the tumor evolves and changes over time and in response to therapy.”
In a new paper, “The tumor microenvironment across four dimensions: assessing space and time in cancer biology,” Carstens and her co-authors explain that the boom in spatial analysis “has exponentially accelerated our understanding of the structural complexity” of the tumor microenvironment. Now, “by studying the tumor microenvironment in the context of space and time (four dimensions), a more complete narrative of each cancer can be written. Patients could be better classified into responders and non-responders for specific therapies. Scientists could identify when and where a target is most vulnerable to manipulation and which cells are nearby to assist in the targeting. Understanding the spatiotemporal changes of a TME would improve biopsy analysis to advance patient therapy and outcome.

But wrestling conclusions out of the flood of data from a tissue sample, which might contain a million tagged cells, is no easy feat. No two tumor neighborhoods will look exactly the same. Which ones are similar enough to put in a cluster that could offer a pattern for clinicians to target? For help, researchers like Carstens look to algorithms developed by ecologists.
Ecological models, as one paper explains, reveal “nonrandom spatial relationships.” One of the most well known is the Morisita-Horn index, which measures similarity among community structures. It helped ecologists establish a positive association between predator body size and prey diversity. The same index has been applied to breast tumors to quantify colocalization of immune and cancer cells. A high Morisita score means that immune cells and cancer cells are found close together. If a high score is associated with a good prognosis, that might mean that the immune cells are effectively preying on cancer cells. A high Morisita score associated with poor prognosis, on the other hand, could indicate that immune cells are being co-opted by the tumor.
This work is inherently multi-disciplinary. “We collaborate with pathologists, surgeons, endoscopists and oncologists who work on large enough patient cohorts so that we can look at survival differences,” Carstens said. “Being at a comprehensive cancer center really helps. Then we work with quantitative radiologists and bioinformaticians to develop the math algorithms to quantify the immune infiltration.”
Feeding annotated comic strips into a machine-learning model could help doctors predict what will happen next for individual patients. It could also point to the treatments with the best chance of halting cancer spread in the particular patient’s case.
“One thing my bioinformatic collaborators and I are really interested in is whether we can create these temporal analyses in mice and then use AI/machine learning to learn enough to take snapshot data from the clinic and extrapolate it out in time,” Carstens said.

“Think about how we predict the weather,” she said. “We have years of snapshots and how they developed over time so that now with good accuracy we can predict if a hurricane is coming to your house or not. Can we feed these AI models enough information from snapshots of cancer model systems so that we really do have a movie of what is going to happen? Then we could test and refine that system over time to generate useful suggestions for clinicians.”
This is both “far away and not far away,” Carstens said. “This is what I think we will be doing 10 years from now. For my collaborators and me, that is what makes us excited.”
At UAB, Carstens has found a community of researchers interested in spatial analyses and techniques. “There are a lot of people here who do fantastic work,” she said. With funding from the Heersink School of Medicine, Carstens is working with other researchers to develop training courses in spatial biology in order to draw trainees and seasoned investigators alike into the field.
“The tools are getting better all the time,” she said. “And they can be used to address much more than cancer. People working in kidney disease, heart disease, stroke and more are applying spatial biology here at UAB. The community is growing.”
Learn more about Carstens and her research.
Want to get involved with spatial biology?
Carstens notes that there are a number of opportunities at UAB.
- Vendor shows and educational seminars are available throughout the year, as well as Spatial Days highlighting UAB investigators and applications of these technologies at UAB. (Check with the sponsoring Core facilities for details.)
- In January 2026, a Spatial Biology and Bioinformatics Journal Club will debut, open to all interested in the topic and available for graduate school credit.
- New graduate-level curricula are being designed, which will include general-knowledge spatial multiomic techniques and applications, as well as hands-on bioinformatics training. These curricula are expected to roll out in fall 2026 and spring 2027, Carstens says.
- In addition, the Spatial Biology PRIME postdoctoral recruitment and training program began recruitment in fall 2025.